|
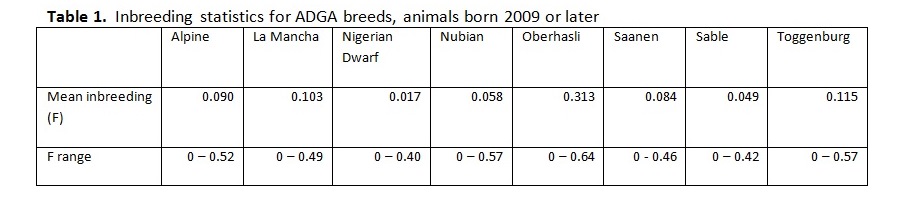
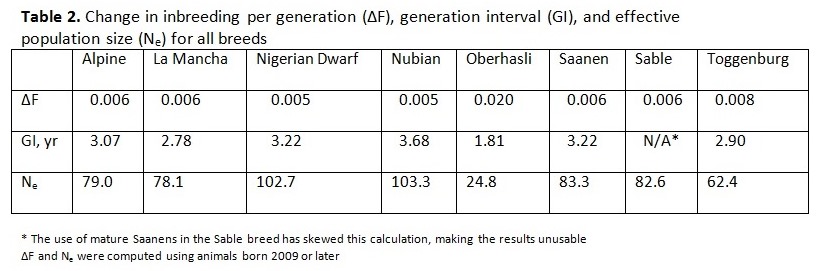
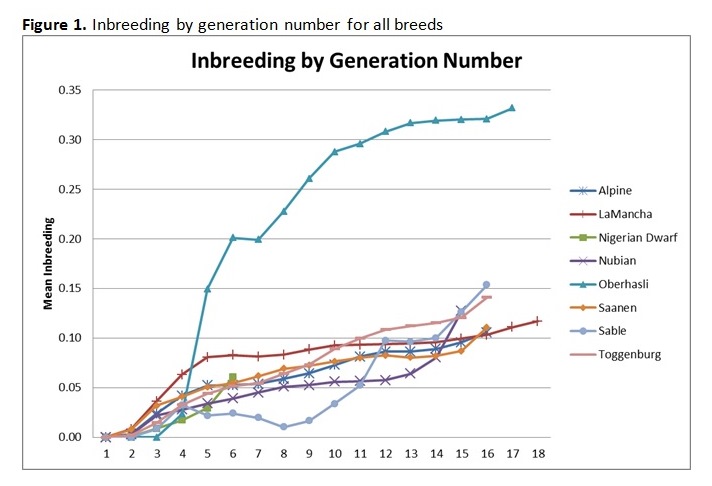
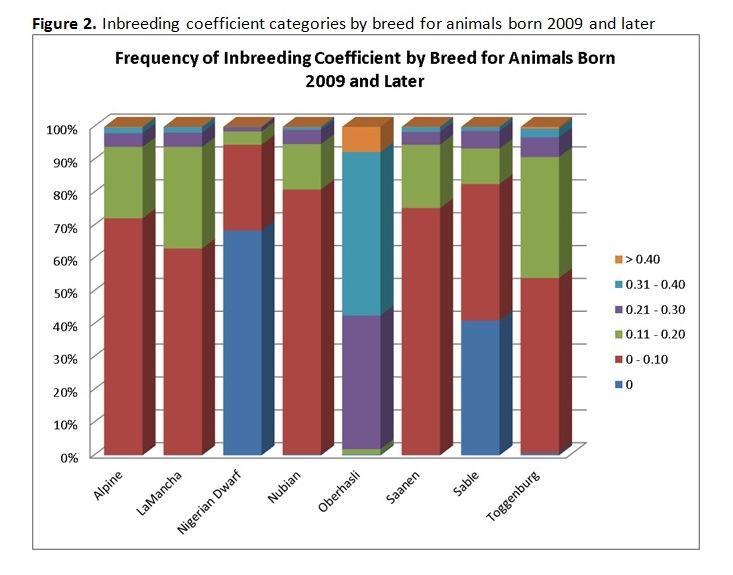
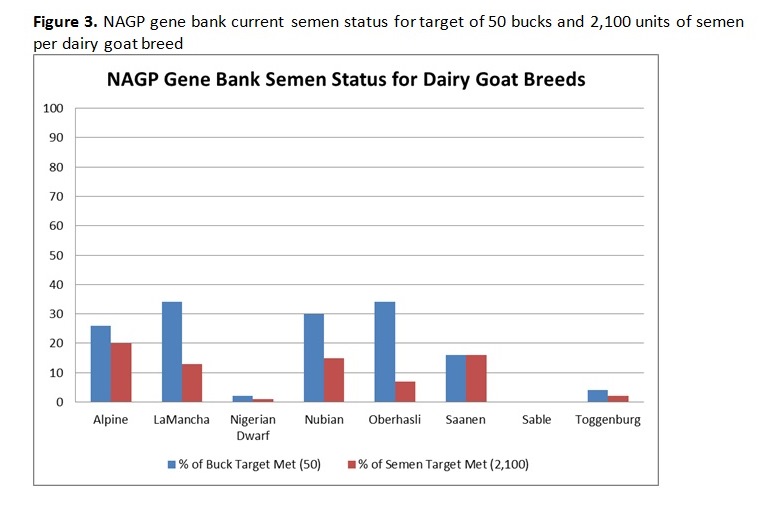
Written by C. S. Wilson, B. Krehbiel, and H. D. Blackburn Genetic diversity underpins the livestock breeders’ ability to improve the production potential of their livestock. Therefore, it is important to periodically assess genetic diversity within a breed. Such an analysis was conducted on U.S. dairy goat breeds and this article is an overview of that work. In March 2014, pedigree files were obtained from the American Dairy Goat Association (ADGA) in order to conduct a genetic diversity evaluation using pedigree information and assess how well the bucks in the National Animal Germplasm Program (NAGP) gene bank represent various goat breeds. NAGP is part of the U.S. Department of Agriculture. The primary function of the gene bank is to serve as a strategic reserve of genetic diversity for the livestock industry to use, particularly in the event of a local or national crisis such as rebuilding breeds after an epidemic. Additional material is in the gene bank for use by breeders and researchers for DNA analysis or reintroduction of genetics that are no longer commercially available. The NAGP gene bank currently includes more than 22,700 animals and 778,000 units of germplasm and various tissues. Of this collection, 409 animals (9,657 units) are goats. Within the goat collection, 146 animals (1,900 units) are from dairy breeds and, within that, 73 animals (1,542 units) have semen stored. Pedigree Evaluation With pedigree information, various measures of genetic relatedness and diversity can be computed. For example, the genetic relationship between a parent and offspring is 0.5 since half of the offspring’s genes came from each parent. Full siblings also have half of their genes in common by descent and a relationship coefficient of 0.5, while half siblings have a relationship coefficient of 0.25. While these computations are basic, full pedigrees with many generations result in complex relationship values best computed by software programs designed to do so. Knowing the pedigree of animals also allows us to compute the inbreeding coefficient for each animal. How this average inbreeding level changes per unit of time for the breed is referred to as the rate of inbreeding (ΔF). Since each breed has its own unique start date, which impacts inbreeding levels over time, comparing breeds based on birth year lacks usefulness. Therefore, animals were assigned a generation number so breeds could be compared on the same basis. Founder animals are defined as having both parents unknown and were assigned to generation zero. Then, subsequent generations were calculated as: g = 1/2 (gs + gd) + 1, where gs is the generation number of the sire and gd is the generation number of the dam. Effective population size (Ne) is defined as the number of individuals that would generate the current rate of inbreeding in a randomly mated population. While there are several ways to compute effective population size, we selected and used the following formula: Ne = 1/(2ΔF) Generation intervals (GI) were computed by regressing generation number on birth year. Pedigree Analysis Results Breed specific results are available on the NAGP website at: http://ars.usda.gov/Main/docs.htm?docid=23140. Between breed results are summarized here. Table 1 shows the inbreeding statistics for each breed. All results were limited to those animals reported as Purebred (PB) or American (AM); however, all animals were included in the pedigree analysis to establish ties between animals, including cases where the ancestors are from another breed. The rate of inbreeding drives the Ne. The higher the ΔF, the smaller the Ne. An inbreeding rate less than 0.005 has been suggested by one researcher (Nicholas, 1989), which equates to an Ne of 100, while the Food and Agricultural Organization of the United Nations (FAO, 2000) suggests a goal of less than 0.01, where 0.01 equates to an Ne of 50. Seven of the eight dairy goat breeds are within the sustainable limits for ΔF and Ne (Table 2). When analyzing several breeds that serve a similar function, it is interesting to make comparisons between them. When inbreeding for all breeds is compared on a per generation basis, the trend is a small to moderate increase over time for all breeds except Sable and Oberhasli (Figure 1). Since Sables have an open herd book breeders can introduce genetic variability from Saanen – as a result they can easily manage inbreeding problems. Historical reports suggest the Oberhasli breed may have been based on few founder animals, which has contributed to higher inbreeding level than the other breeds. Seven of the eight breeds have data at 15 generations; the range of inbreeding at generation 15 is 0.087 for Saanen to 0.320 for Oberhasli. The next highest inbreeding level at generation 15 is Nubian with 0.127. Our analysis suggests that 99% of current animals among breeds have an inbreeding coefficient greater than zero. The generation number the breeds reached this level was: generation 7 for Alpine, Nigerian Dwarf, and Saanen; generation 8 for Nubian and Toggenburg; generation 9 for Lamancha and Oberhasli; and generation 12 for Sable. Figure 2 compares inbreeding categories for the current population (animals born 2009 and later). There is a large difference across the breeds, with Nigerian Dwarf having the most non-inbred animals and Oberhasli having the highest percentage of highly inbred animals. There are very few lowly related animals that can be used to reduce the inbreeding level within the Oberhasli breed. Using a buck to doe ratio close to 1.0 is a strategy that can be used to widen the genetic base and keep inbreeding levels lower than allowing a few males to dominate the breed. Nigerian Dwarfs have a shallower pedigree than the other dairy goat breeds. Computationally, inbreeding has not had much time to accumulate, leading to low inbreeding levels for this breed. The inbreeding levels are probably higher in reality since inbreeding was accumulating as the breed was established rather than as the herdbook was established. Molecular analysis to better establish a baseline inbreeding level would be particularly useful for this breed. Because Sable has an open herdbook with the ability to use unrelated Saanens, breeders can easily introduce unrelated genetics to manage inbreeding levels. Implications for Breeders and NAGP Inbreeding can be part of a mating plan to establish or maintain characteristics of interest within a line, for example. Alternatively, and far more prevalent, is incidental inbreeding that happens over time when lowly related animals are mated and inbreeding slowly accumulates until all animals have some level of inbreeding. In the case of planned linebreeding, inbreeding can be used to fix (make homozygous) alleles for a trait, leading to a more uniform phenotype. All inbreeding, whether intentional or incidental, leads to more homozygosity and, eventually, a potential loss of lower frequency alleles and thereby a loss of genetic diversity. The increase in homozygosity with inbreeding can also bring out deleterious recessives that were previously hidden in a population; this is known as inbreeding depression and can lead to reduced performance. Purging deleterious recessives is part of establishing an inbred line. While a uniform phenotype is desirable to the breeder and consumer, it also limits the ability to respond to market changes if the formerly undesirable alleles become preferred. Breeders can use pedigree analysis and inbreeding calculations as tools to plan matings and predict inbreeding for their herd for future generations. One such tool is available on the ADGA website at: http://www.adgagenetics.org/Default.aspx. This excellent tool allows breeders to select a potential buck and doe for mating to see the inbreeding coefficient, pedigree, and Estimated Transmitting Ability of the offspring. Developing a genetically diverse collection of dairy goat semen requires access to dairy goats for collection and/or semen contributions of previously collected bucks. Because artificial insemination is not as commercially established as with the dairy cattle industry, greater effort is required on the part of NAGP to obtain the samples needed for the gene bank. Figure 3 shows the status of dairy breed collections in the NAGP gene bank as a percent of the targeted germplasm and number of bucks collected. The NAGP dairy goat collection is short of the targets for capturing genetic diversity across breeds, but this can be quickly reversed. Expansion of the dairy goat collection is necessary to develop a reserve of genetic diversity for worst case scenarios. We therefore wish to encourage breeders to work with us in identify existing cryopreserved semen or from bucks that may be collected in the near future to add to the gene bank. Information about NAGP can be obtained at: http://www.ars.usda.gov/main/site_main.htm?modecode=30-12-05-00. Donations can be arranged by contacting Carrie Wilson at [email protected]. Citations
FAO. 2000. Secondary guidelines for development of national farm animal genetic resources management plans: management of small populations at risk. UN Food and Agric. Org. http://www.fao.org/AG/AGAInfo/resources/en/pubs_gen.html. Accessed March 12, 2009. Nicholas, F. W. 1989. Incorporation of new reproductive technology in genetic improvement programmes. Pages 201-209 in Evolution and Animal Breeding. W. G. Hill and T. F. C. Mackay, ed. CAB Int., Wallingford, U.K.
1 Comment
|
IGA Blog
The International Goat Association promotes goat research and development for the benefit of humankind, to alleviate poverty, to promote prosperity and to improve the quality of life. Archives
May 2024
Categories
All
|
|
International Goat Association
2516 Millbrook Rd., Little Rock, AR72227 USA email: [email protected] -454-1641 |

 RSS Feed
RSS Feed